Using VuTR
Searching for the gene of your choice
You can easily search for a gene of your choice using gene names. We plan to introduce search options through other entities such as Refseq and Ensembl Transcript IDs. To search for a gene of your choice, visit the Home page and enter the gene’s symbol name (e.g. MEF2C). For each gene searched, users are directed to a viewer with the following cards.
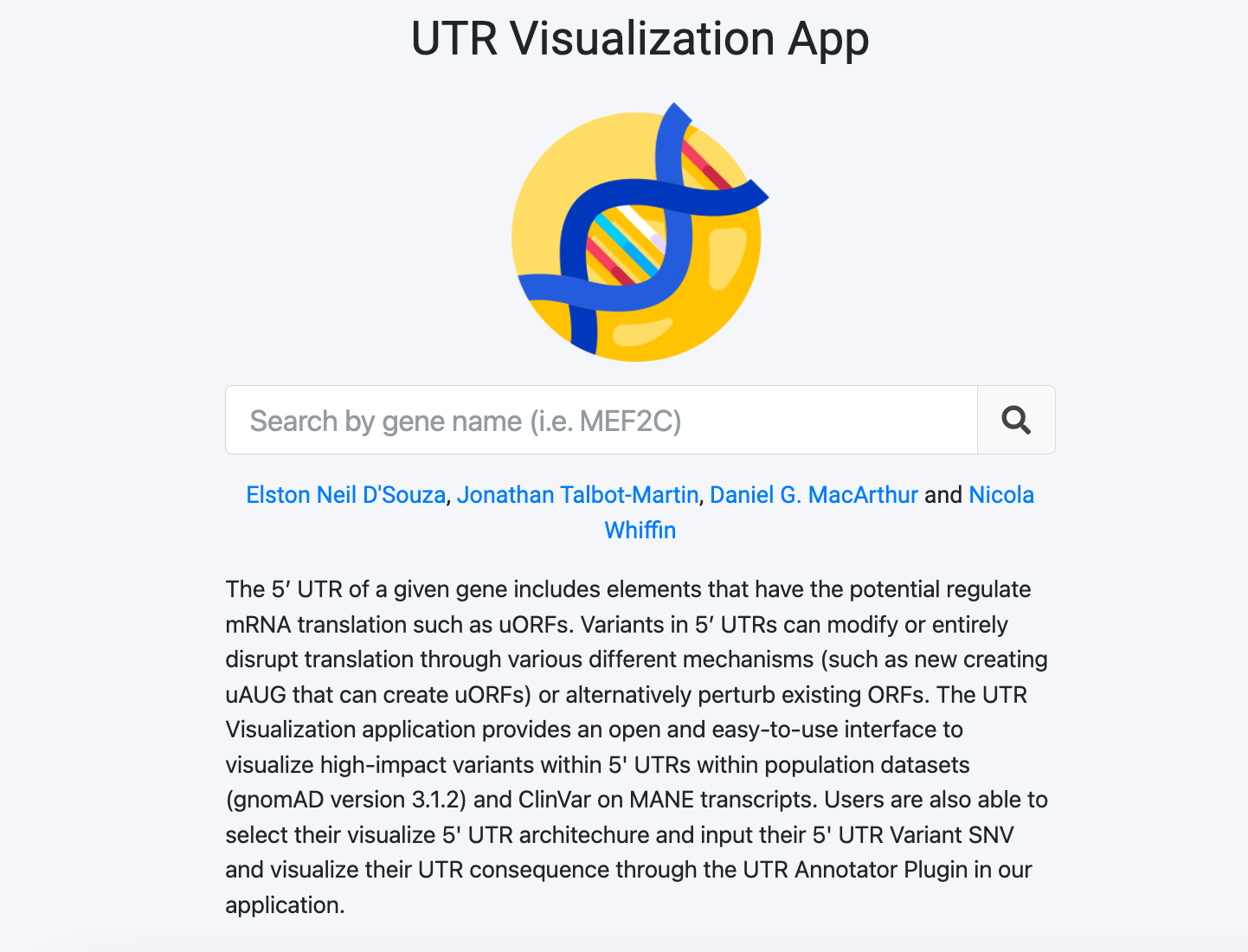
Gene detail card
The gene detail card contains information about the gene chosen including alternate identifiers and links to external sources for that gene. We include the following information:
- HGNC Symbol: The gene's HGNC symbol
- Gene Name: The gene's common name
- Ensembl Gene ID: Ensembl's gene identifier (e.g. ENSG000001000)
- Ensembl Transcript ID: The Ensembl transcript of that gene according to MANE Select 1.0
- RefSeq Match: The RefSeq transcript identifier according to MANE Select 1.0
- 5' UTR exon count:The number of exons in the 5' UTR
- 5' UTR length: The length of the 5' UTR (in bps)
- Gene LOEUF: Constraint scores according to gnomAD v.2.1.1
- External Resources See this gene in other resources such as gnomAD, ClinVar, sORFs, and ClinGen.
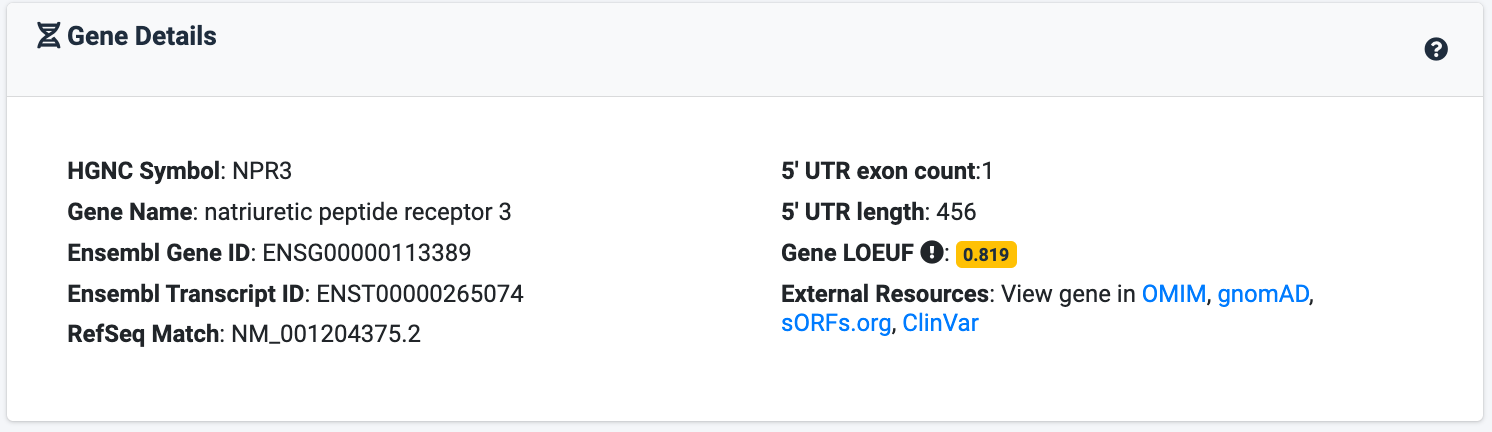
Visualising the 5' UTR Architechure
The UTR Visualisation app is able to visualise the various different uORFs (upstream Open Reading Frames) and oORFs (overlapping Open Reading Frames) within a 5' UTR. This is done by scanning through from the 5' to 3' end of the UTR and identifying all ATGs with an in-frame stop codon. These are then characterised as whether they are (a) a uORF with a stop codon in-frame within the UTR (b) in-frame oORFs causing an N-terminal extension or (c) an out-of-frame oORF. These ORFs are separately viewed on different tracks on the viewer. They are colour coded with their match to Kozak Consensus Sequence (5'-(gcc)gccRccAUGG-3') and categorised as either (a) Strong, (b) Moderate or (c) Weak.
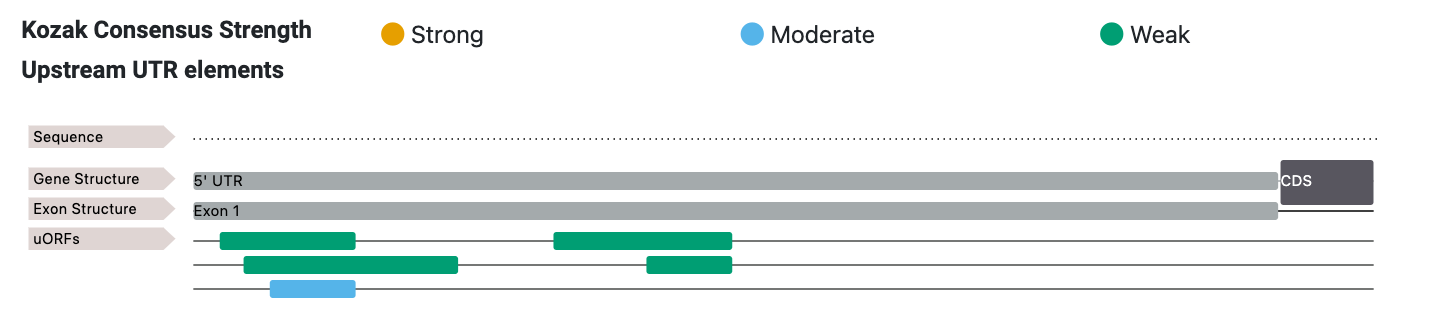
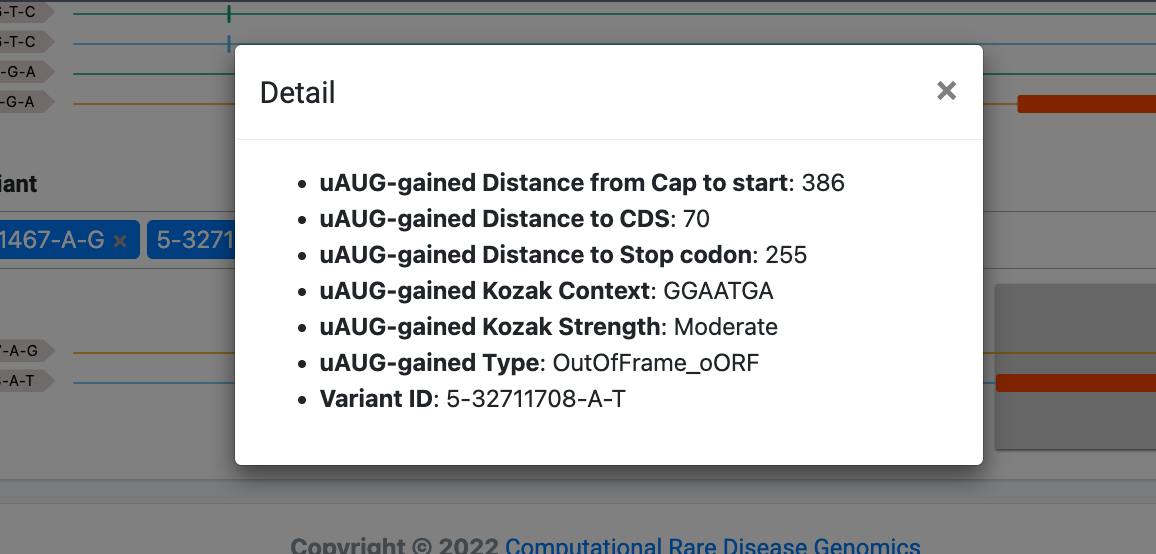
Users are able to view additional contextual detail such as the sequence, frame, context and transcript positions by clicking on the ORF of their choice.
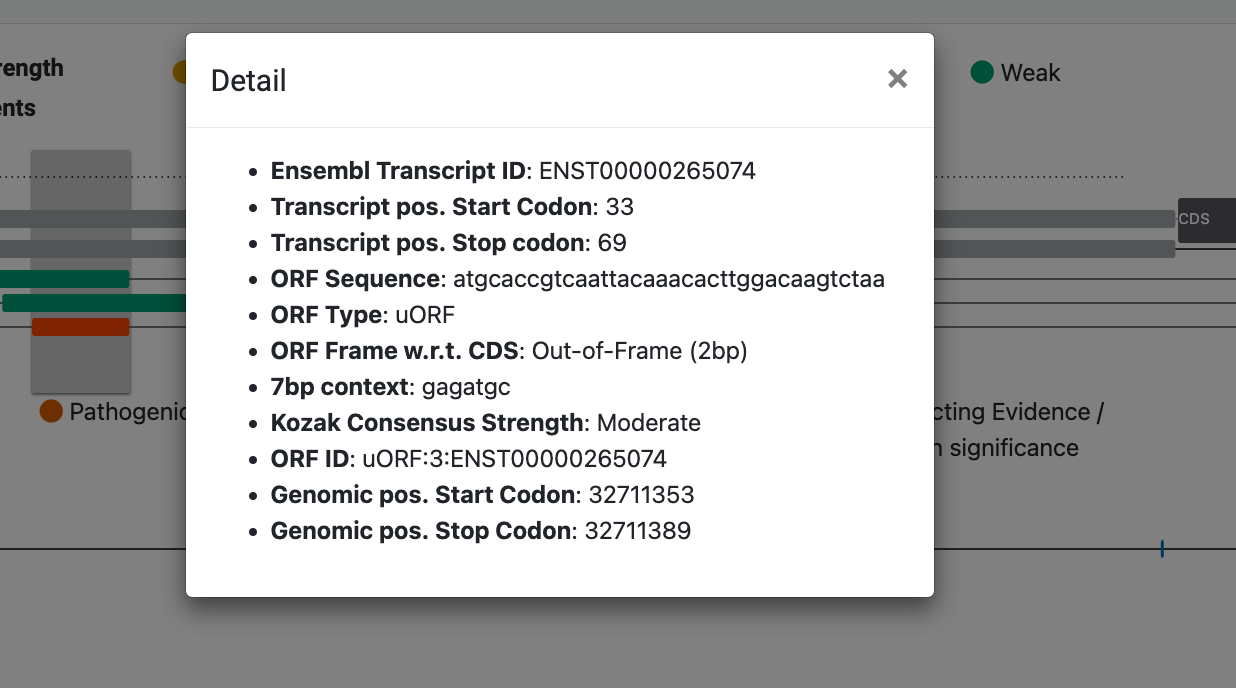
ClinVar and gnomAD variants
We use gnomAD's GraphQL API to access variants within 5' UTR exons. ClinVar variants are placed in separate tracks, one for each variant with a 5' UTR consequence. gnomAD (version 3.1.2) was used for a population reference variation.
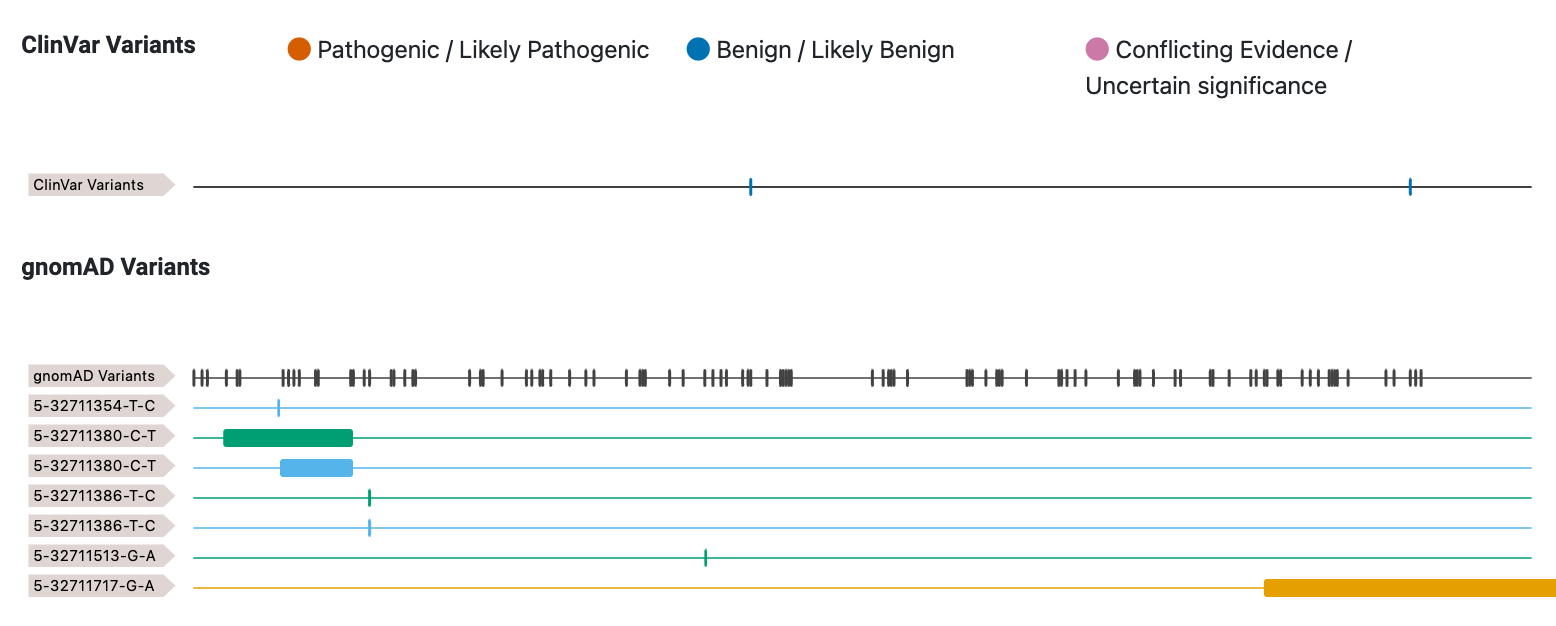
Click on the track to get more contextual details on the specific variant.
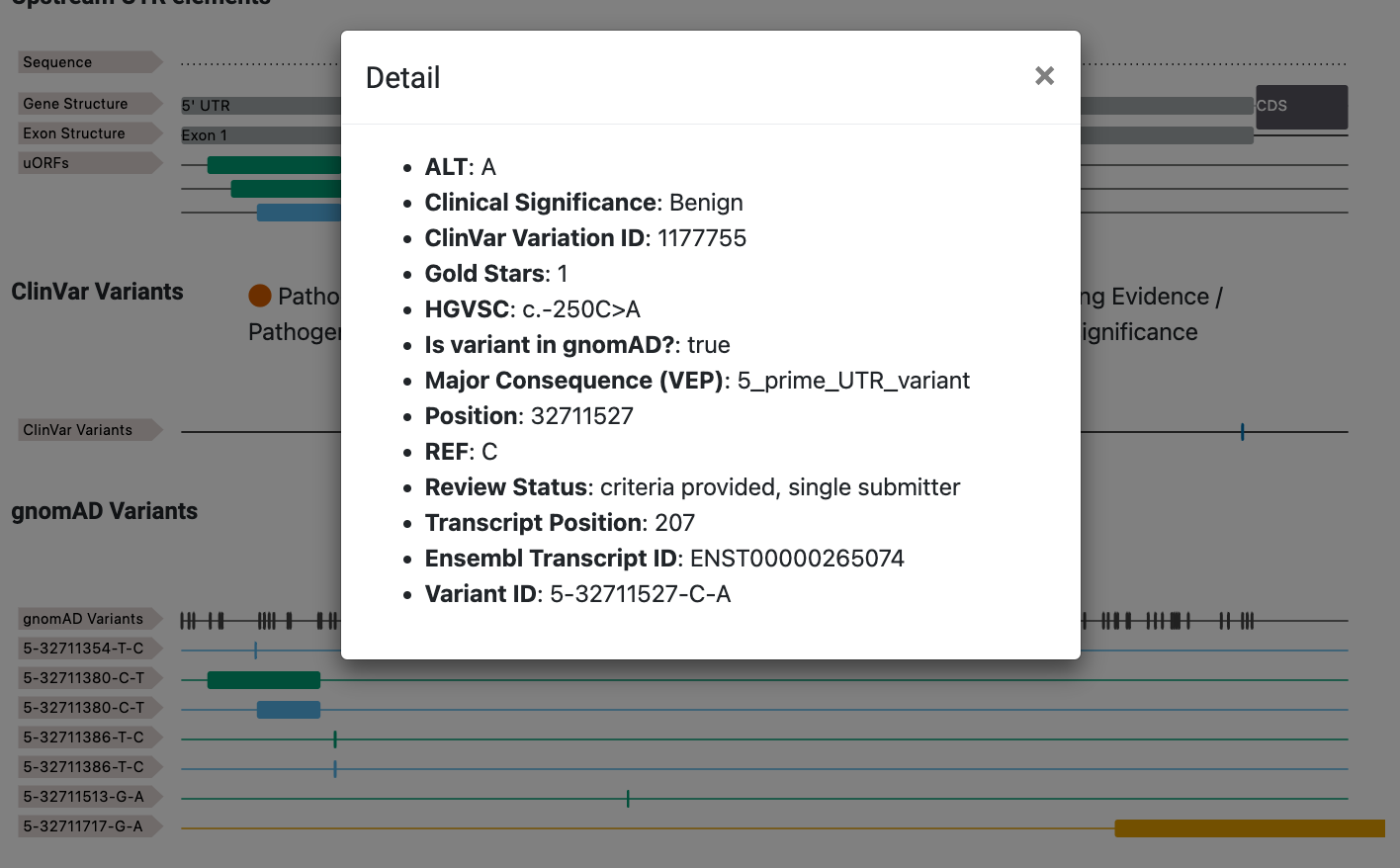
Finding the impact your favourite variant
Users are available to input their favourite variant in the box user-supplied variant section in the following format CHROM-POS-REF-ALT (e.g. 5-88823796-G-A). Only variants with a 5' UTR impact are searchable, all other SNV variants in 5' UTR regions are assumed to not create or disrupt an existing uORF or oORF.

Clicking on the ORF that is either created or gain will allow users to view the consequences provided from the VEP Plugin UTR annotator.
Frequently Asked Questions (FAQs)
Can I get a copy of the data?
Please get in touch with us regarding data access. We are working to making our data as openly as possible. Until then, get in touch with us here
What caveats do I need to look out for whilst using the UTR annotator?
The UTR application (a) currently uses SNVs although we have plans to include small (less than 3bp) indels in our application (b) intronic variants such that impact splicing can perturb the UTR architechure however the UTR Annotator doesn't support intronic variants yet.
Who do I contact regarding an error or a bug?
If you face any issues or wish to report a bug whilst using the UTR Visualisation Application please get in touch with us here or by submitting a ticket through our Github issues page.